A novel methodology on distributed representations of proteins using their interacting ligands
The effective representation of proteins is a crucial task that directly affects the performance of many bioinformatics problems. Related proteins usually bind to similar ligands. Chemical characteristics of ligands are known to capture the functional and mechanistic properties of proteins suggesting that a ligand based approach can be utilized in protein representation.
SMILESVec, a SMILES-based method to represent ligands and a novel method to compute similarity of proteins by describing them based on their ligands. The proteins are defined utilizing the word-embeddings of the SMILES strings of their ligands.
We showed that ligand-based protein representation, which uses only SMILES strings of the ligands that proteins bind to, performs as well as protein-sequence based representation methods in protein clustering. The results suggest that ligand-based protein description can be an alternative to the traditional sequence or structure based representation of proteins and this novel approach can be applied to different bioinformatics problems such as prediction of new protein-ligand interactions and protein function annotation.
Representation of Proteins
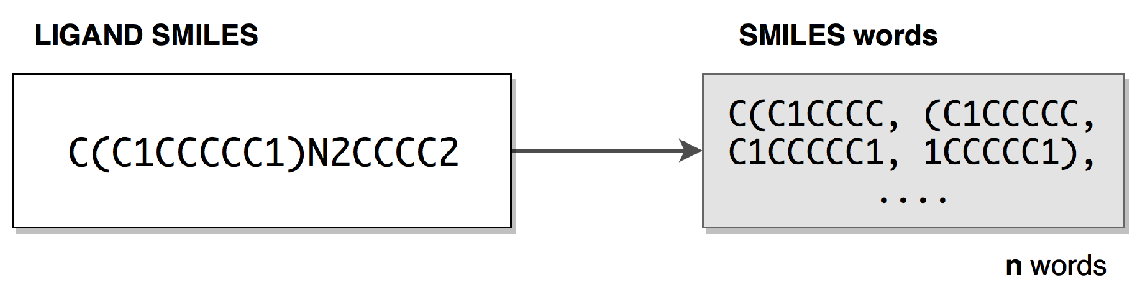
Figure above explains the construction of chemical words from an example SMILES string. The chemical words are eight-charactered overlapping subsequences. For each subsequence, a continous vector is assigned which was created via Word2Vec model after learning on a large SMILES corpus. As SMILES corpus, we utilized ChEMBL and PubChem .
SMILESVec, the ligand representation vector, is calculated as the average of the n chemical word (subsequence) vectors that are created from SMILES string.
Publication
Öztürk, Hakime, Elif Ozkirimli, and Arzucan Özgür. "A novel methodology on distributed representations of proteins using their interacting ligands."
Bioinformatics, accepted for publication (2018).
ISMB2018 Proceedings | arXiv
Bioinformatics, accepted for publication (2018).
ISMB2018 Proceedings | arXiv
Embedding Data
| File | Description |
|---|---|
| drug.l8.chembl23.canon.ws20 | word-level embeddings trained on ChEMBL23 |
| drug.l8.pubchem.canon.ws20 | word-level embeddings trained on Pubchem |
| drug.l1.chembl23.canon.ws20 | char-level embeddings trained on ChEMBL23 |
| drug.l1.pubchem.canon.ws20 | char-level embeddings trained on Pubchem |
Source Code
Implementation of the method and SCOP A-50 data set that we used for evaluation are available on Github.