Ligand-centric network models
PLITOOL provides ligand-centric networks as outputs that are explained below, namely: identity networks and similarity networks.
The studies on biological networks enable better understanding of the mechanism of interaction between molecules. Our studies revolve around protein-protein interaction networks which are created using the ligands that they bind to.
We build two different types of networks in which the proteins are represented as nodes, and two proteins are connected by an edge with a weight that depends on the number of shared identical or similar ligands. These models are analyzed under three different edge weight settings, namely unweighted, weighted, and normalized weighted.
A detailed comparison of these six networks showed that the use of ligand sharing information to cluster proteins resulted in modules comprising proteins with not only sequence similarity but also functional similarity. Consideration of ligand similarity highlighted some interactions that were not detected in the identical ligand network. Analysing the β-lactamases and PBPs using ligand-centric network models, that we experimented on as a case study, enabled the identification of novel relationships, suggesting that these models can be used to examine other protein families to obtain information on their ligand induced evolutionary paths.
Identity Networks
The identity network model is based on sharing of common ligands. In this model two proteins are connected with an edge if they share at least one identical ligand.
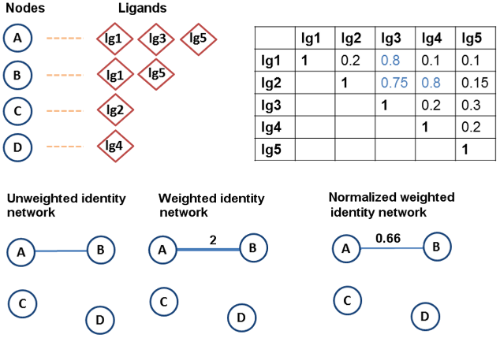
A sample data set consisting of four proteins (A, B, C, D) shaped as circles and five ligands (lg1, lg2, lg3, lg4, lg5) shaped as diamonds. For each protein, the ligands that it binds to are given together. A sample Tanimoto coefficient (Tc) matrix is also provided for the ligand pairs. (The same example is used in the next figure.) In the identity networks, A and B are connected since they have two common ligands, lg1 and lg5. Only the weight of the edge between A and B changes depending on the weighting method used.
Similarity Networks
This model enables us to link two nodes that do not have any common ligands, but bind to ligands whose chemical similarity is above some pre-determined threshold. In this study, the similarity threshold was selected as Tc of 0.7. In other words, if two nodes shared two ligands with more than 0.7 Tc, they were connected in the similarity network. The similarity based model aims to discover some hidden relationships or emphasize the existing ones using the ligand chemical similarity feature.
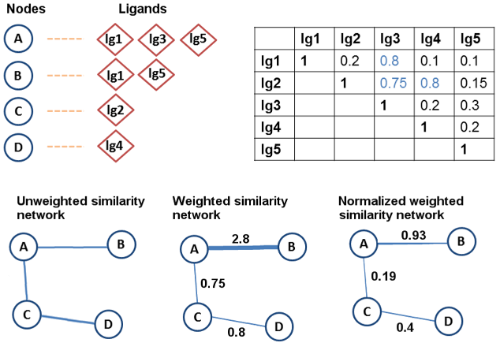
In the similarity networks, the proteins that bind to ligands whose pairwise similarities exceed the Tc 0.7 cut-off value are connected. Therefore, we have two new connections in the similarity network: C and D are connected due to the similarity between lg2 and lg4, and A and C are connected due to the similarity between lg2 and lg3. The edge weights between nodes change depending on the weighting method used.
Publication
Öztürk, Hakime, Elif Ozkirimli, and Arzucan Özgür. "Classification of Beta-lactamases and penicillin binding proteins using ligand-centric network models.".
PloS one, 2015
Plos One | Bibtex
PloS one, 2015
Plos One | Bibtex
Source Code
| File | Description |
|---|---|
| ligandcentricnetworkv3.jar | jar to run ligand-centric models |
| drug.l8.chembl23.canon.ws20 | SMILESVec word-level embeddings trained on ChEMBL23 |
| input.txt | Example input file |
| ide_unwei.txt | Example output file (unweighted identity network) |
| README.txt | README file |